Kennard-Stone (KS) and SPXY algorithms are common sample split algorithms in the chemometrics field. Unlike random split, these two methods are based on the so called "maximum minimum distance", which means the split result of KS and SPXY are definite instead of random. This article will briefly introduce these 2 algorithms and provide the python implementation.
Background
Multivariate data analysis is a pillar of chemometrics, and multi-dimensional data are quite easily acquired in the presence of new instruments, such as spectrometers. Multi-dimensional data should be carefully split into train, validation, and test set to build chemometrics models. Random split is a common choice, but the models can vary due to different split results. What's worse, random split can't assure a proper split result, i.e., the subsets after split can't not represent the original dataset. So many researches have investigated how to select a representative subset from a large dataset.
KS and SPXY algorithms are commonly used in spectra analysis. The 2 methods can select samples uniformly from a pool of samples. KS algorithm are conducted based on the similarity of independent variables () between the subset and the original set, while SPXY combines independent and dependent variables ()[1]. They are very similar, both selecting subset samples by maximum minimum distance.
KS split
Assume we are going to select samples to create a subset from samples. The samples are presented by the matrix .
where is the number of variables of each sample.
The steps of KS split is shown below:
- Calculate the distance matrix of the samples as the similarity matrix. Euclidean distance is often adopted here. The distance matrix is denoted as below.
where is the distance between the th and the th sample. So is a symmetric matrix if the distance is Euclidean distance.
- Add the 2 samples between which the distance is the longest into the subset . Now we face the maximum minimum distance problem. There are 2 samples in (named and , respectively) and -2 samples remaining now.
- Select a sample c from remaining samples, calculate the distance of c to and , respectively (You don't have to calculate again indeed, because all distances have been calculated in step1).
- If the distance between and is shorter than the distance between and , then we call the distance "minimum distance of sample ".
- Repeat the step 3 and step 4 above, then we can get all the minimum distances of the remaining -2 samples.
- From the -2 minimum distances, the maximum distance (maximum minimum distance) is selected as the new sample of , shown in the figure below.
- Repeat the steps above until samples are added into .
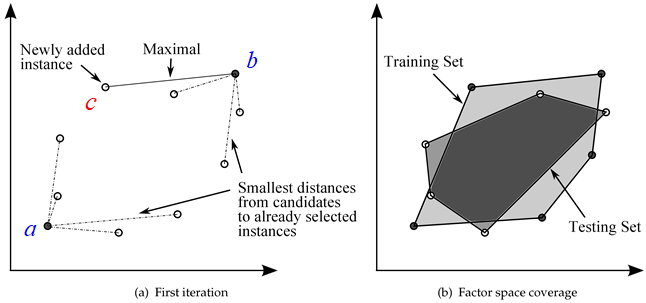
The samples in are distributed uniformly and by this method, the train set will represent the original set to some extent.
SPXY split
As illustrated at the start, KS split only concerns the similarity (distance) between independent variables, while SPXY adds dependent variables into distance calculation. The only difference between KS and SPXY is the way to calculate distance. Assume we have sample matrix is shown as
The distance calculation formula for KS and SPXY are shown below:
In fact, the core of KS and SPXY algorithms are maximum minimum distance split, and we can define another distance metric according to the real situation. A research proposed a new distance metric based on cosine similarity[2].
Python implementation
# -*- coding=utf-8 -*-
from __future__ import division, print_function
import numpy as np
from sklearn.model_selection import train_test_split
from scipy.spatial.distance import cdist
def random_split(spectra, test_size=0.25, random_state=None, shuffle=True, stratify=None):
"""implement random_split by using sklearn.model_selection.train_test_split function. See
http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html
for more infomation.
"""
return train_test_split(
spectra,
test_size=test_size,
random_state=random_state,
shuffle=shuffle,
stratify=stratify)
def kennardstone(spectra, test_size=0.25, metric='euclidean', *args, **kwargs):
"""Kennard Stone Sample Split method
Parameters
----------
spectra: ndarray, shape of i x j
i spectrums and j variables (wavelength/wavenumber/ramam shift and so on)
test_size : float, int
if float, then round(i x (1-test_size)) spectrums are selected as test data, by default 0.25
if int, then test_size is directly used as test data size
metric : str, optional
The distance metric to use, by default 'euclidean'
See scipy.spatial.distance.cdist for more infomation
Returns
-------
select_pts: list
index of selected spetrums as train data, index is zero based
remaining_pts: list
index of remaining spectrums as test data, index is zero based
References
--------
Kennard, R. W., & Stone, L. A. (1969). Computer aided design of experiments.
Technometrics, 11(1), 137-148. (https://www.jstor.org/stable/1266770)
"""
if test_size < 1:
train_size = round(spectra.shape[0] * (1 - test_size))
else:
train_size = spectra.shape[0] - round(test_size)
if train_size > 2:
distance = cdist(spectra, spectra, metric=metric, *args, **kwargs)
select_pts, remaining_pts = max_min_distance_split(distance, train_size)
else:
raise ValueError("train sample size should be at least 2")
return select_pts, remaining_pts
def spxy(spectra, yvalues, test_size=0.25, metric='euclidean', *args, **kwargs):
"""SPXY Sample Split method
Parameters
----------
spectra: ndarray, shape of i x j
i spectrums and j variables (wavelength/wavenumber/ramam shift and so on)
test_size : float, int
if float, then round(i x (1-test_size)) spectrums are selected as test data, by default 0.25
if int, then test_size is directly used as test data size
metric : str, optional
The distance metric to use, by default 'euclidean'
See scipy.spatial.distance.cdist for more infomation
Returns
-------
select_pts: list
index of selected spetrums as train data, index is zero based
remaining_pts: list
index of remaining spectrums as test data, index is zero based
References
---------
Galvao et al. (2005). A method for calibration and validation subset partitioning.
Talanta, 67(4), 736-740. (https://www.sciencedirect.com/science/article/pii/S003991400500192X)
"""
if test_size < 1:
train_size = round(spectra.shape[0] * (1 - test_size))
else:
train_size = spectra.shape[0] - round(test_size)
if train_size > 2:
yvalues = yvalues.reshape(yvalues.shape[0], -1)
distance_spectra = cdist(spectra, spectra, metric=metric, *args, **kwargs)
distance_y = cdist(yvalues, yvalues, metric=metric, *args, **kwargs)
distance_spectra = distance_spectra / distance_spectra.max()
distance_y = distance_y / distance_y.max()
distance = distance_spectra + distance_y
select_pts, remaining_pts = max_min_distance_split(distance, train_size)
else:
raise ValueError("train sample size should be at least 2")
return select_pts, remaining_pts
def max_min_distance_split(distance, train_size):
"""sample set split method based on maximun minimun distance, which is the core of Kennard Stone
method
Parameters
----------
distance : distance matrix
semi-positive real symmetric matrix of a certain distance metric
train_size : train data sample size
should be greater than 2
Returns
-------
select_pts: list
index of selected spetrums as train data, index is zero-based
remaining_pts: list
index of remaining spectrums as test data, index is zero-based
"""
select_pts = []
remaining_pts = [x for x in range(distance.shape[0])]
# first select 2 farthest points
first_2pts = np.unravel_index(np.argmax(distance), distance.shape)
select_pts.append(first_2pts[0])
select_pts.append(first_2pts[1])
# remove the first 2 points from the remaining list
remaining_pts.remove(first_2pts[0])
remaining_pts.remove(first_2pts[1])
for i in range(train_size - 2):
# find the maximum minimum distance
select_distance = distance[select_pts, :]
min_distance = select_distance[:, remaining_pts]
min_distance = np.min(min_distance, axis=0)
max_min_distance = np.max(min_distance)
# select the first point (in case that several distances are the same, choose the first one)
points = np.argwhere(select_distance == max_min_distance)[:, 1].tolist()
for point in points:
if point in select_pts:
pass
else:
select_pts.append(point)
remaining_pts.remove(point)
break
return select_pts, remaining_pts